Microbiome
Improve microbial genome coverage and increase detection sensitivity of low abundant microbial species with CRISPRclean.
The human body serves as a host to a community of microorganisms that far outnumber the body’s own cells. The human microbiome plays a critical role in human health. Deciphering the composition and function of the human microbiome can provide a deeper understanding of its structural and functional properties. Our understanding of the human microbiome and the application of metagenomic analyses will greatly enhance our understanding of human health and disease in specific individuals.
Human microbiome sample types are diverse and come from a variety of body sites including saliva, gut, skin, wound infections and everywhere a microbial community can be found. Samples often consist of a mixture of human and microbial cells. For some samples like stool, the composition is generally made up of mostly bacterial species and very little human cells, but nasal swabs is the opposite. One commonly used method for human microbiome research is shotgun metagenomics sequencing, which is used to characterize the complete bacterial phylogeny and taxonomy of complex, microbiome samples. Depending on the sample type and the research goal, human-derived complex samples can be a challenge due to the overwhelming human host contamination that obscures sequencing coverage of interesting microbial genomes.
With CRISPRclean, you can gain an accurate taxonomic profile of a microbial community by removing human host DNA or RNA.
Boost microbial detection with unbiased human DNA depletion
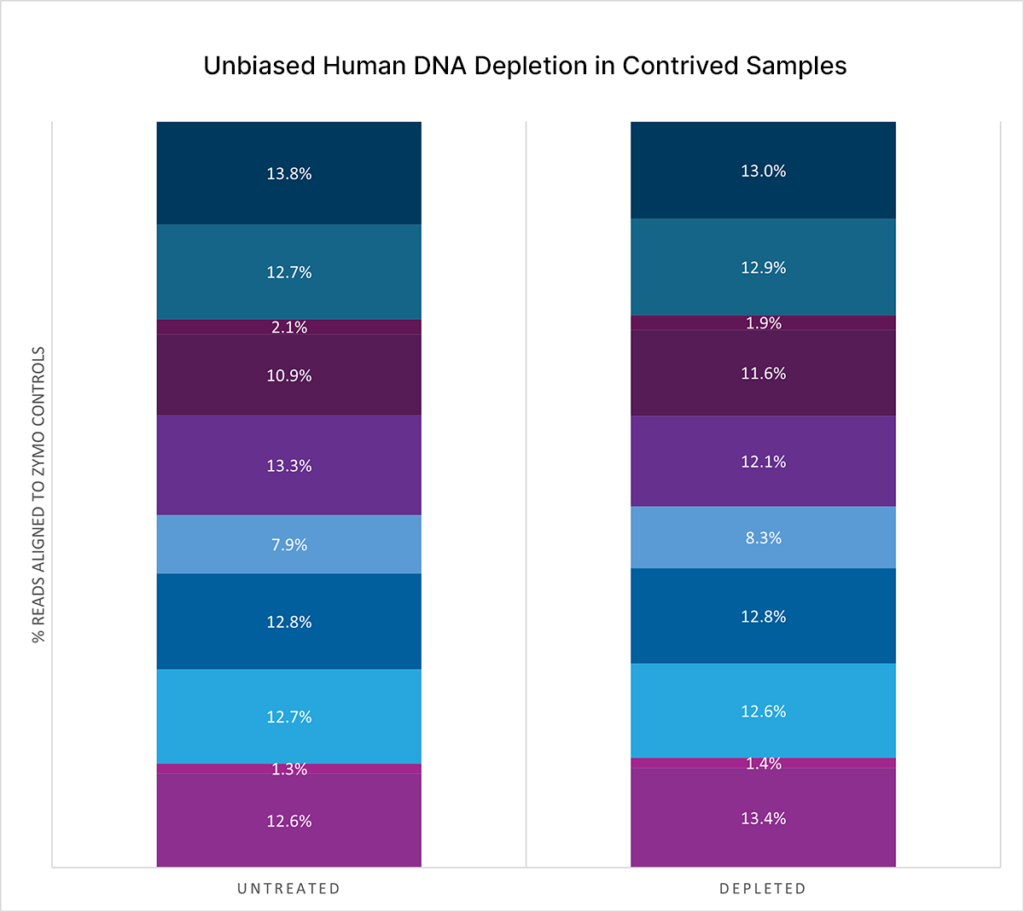
Comparing between untreated and depleted contrived samples, human host depletion preserves microbial composition in the sample.
Depletion increases bacterial detection sensitivity by 4-5 fold
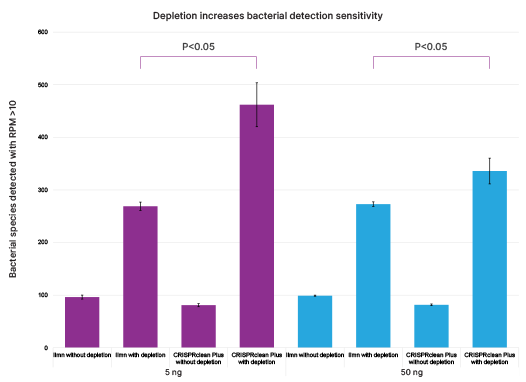
Human rRNA depletion for metatranscriptomics increases bacterial detection sensitivity compared to other methods.
Explore featured resources
CRISPRclean Plus Stranded Total RNA Prep with rRNA Depletion
Increase bacterial species detection in fecal samples
Human DNA Depletion Kit
Connect with us to learn more
For the US patent, the patent number is US 10,604,802 entitled Genome Fractioning. The patent publication is available here.
For the EP patent, the patent number is EP3102722 entitled Genome Fractioning. The patent publication is available here.