
CRISPRclean Plus Stranded Total RNA Prep with rRNA Depletion (Human, Mouse, Rat, Pan Bacteria)
Identify lower expressing transcripts with CRISPRclean Plus
Complex samples may have a mixture of bacterial and human cells, all actively expressing ribosomal RNA. Take advantage of the power of CRISPRclean for effective ribosomal RNA removal. CRISPRclean can also remove uninformative bacterial reads in-vitro ahead of sequencing – redistributing >80% of sequencing reads to enable identification of lower expressing, biologically relevant transcripts.
Highlights
- Streamlined CRISPRclean workflow: stranded total RNA library prep with ribodepletion
- Ideal for analysis of samples containing complex mixtures of eukaryotic and bacterial rRNA
- 3 to 7-fold increase in coverage of SARS-CoV-2 and other genomes in nasopharyngeal (NSP) samples
- Single workflow to detect viral genomic data, microbiome composition, co-infections, and host gene expression
| Specification | |
| Catalog | KIT1016 |
|---|---|
| Samples per kit | 24 samples |
| Assay time | 9 hours |
| Hands-on time | 3.5 hours |
| Sample types | Complex samples with mixtures of eukaryotes and prokaryotes including nasopharyngeal, saliva, gut, skin, and fecal. |
| Input quantity | 5ng - 100ng |
| Strand specificity | >98% directional and strand specific |
| Compatible species | Human, mouse, rat, 212 bacteria |
| Designed to deplete | Human 5S, 5.8S, 18S and 28S, 45S rRNA precursor, mitochondrial 12S and 16S rRNA genes. 212 bacteria representing all phyla: 5S, 16S, and 23S rRNA gene. |
| Multiplex | Up to 96 unique dual index adapter barcodes. Requires KIT1017 CRISPRclean Unique Dual Index Adapter Plate for RNA Prep (Set A) |
CRISPRclean Plus Workflow


Performance
>80% depletion of eukaryotic and bacterial rRNA in clinical NSP samples
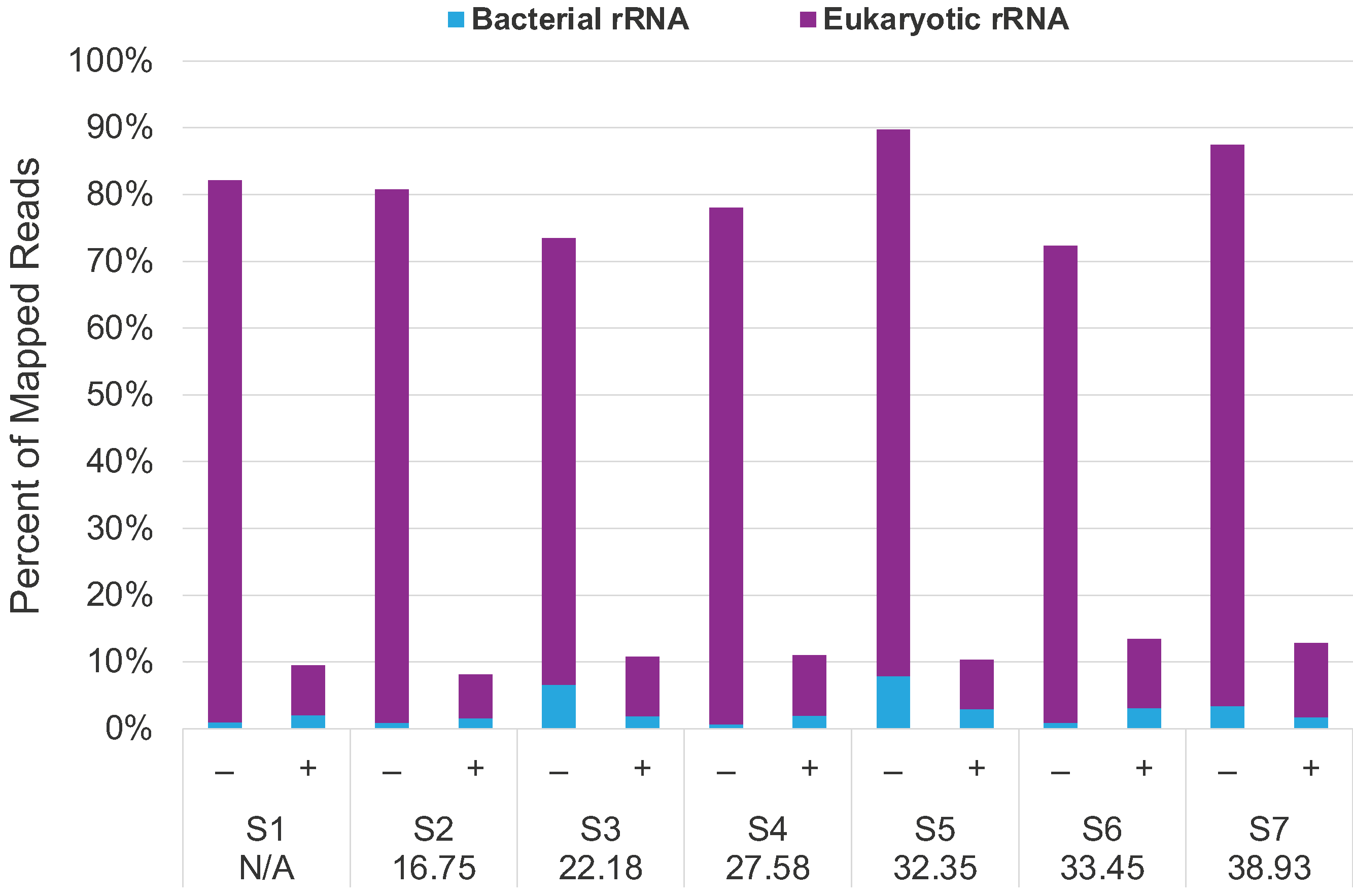
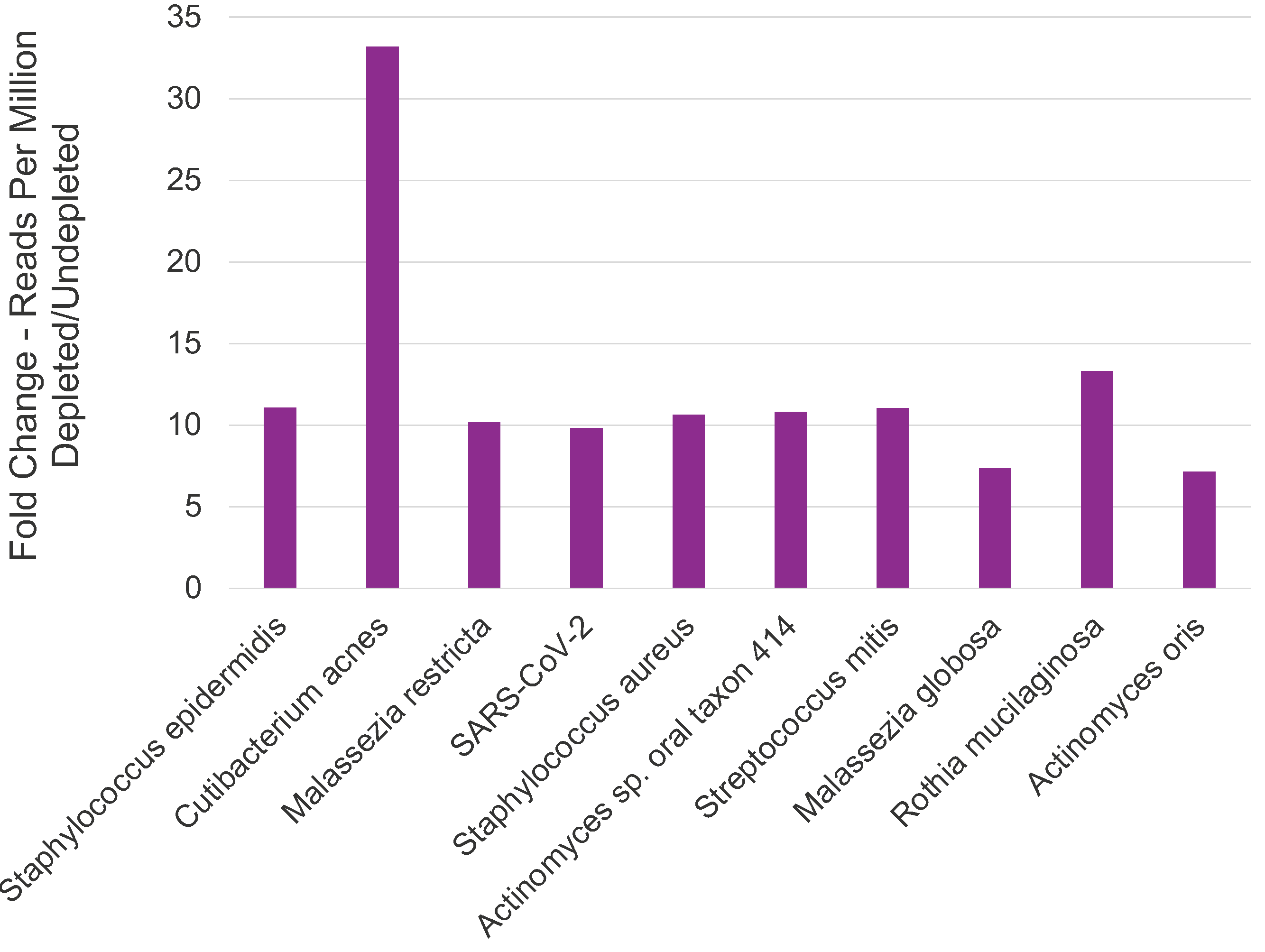
Increased sequencing reads for microbial organisms after depletion
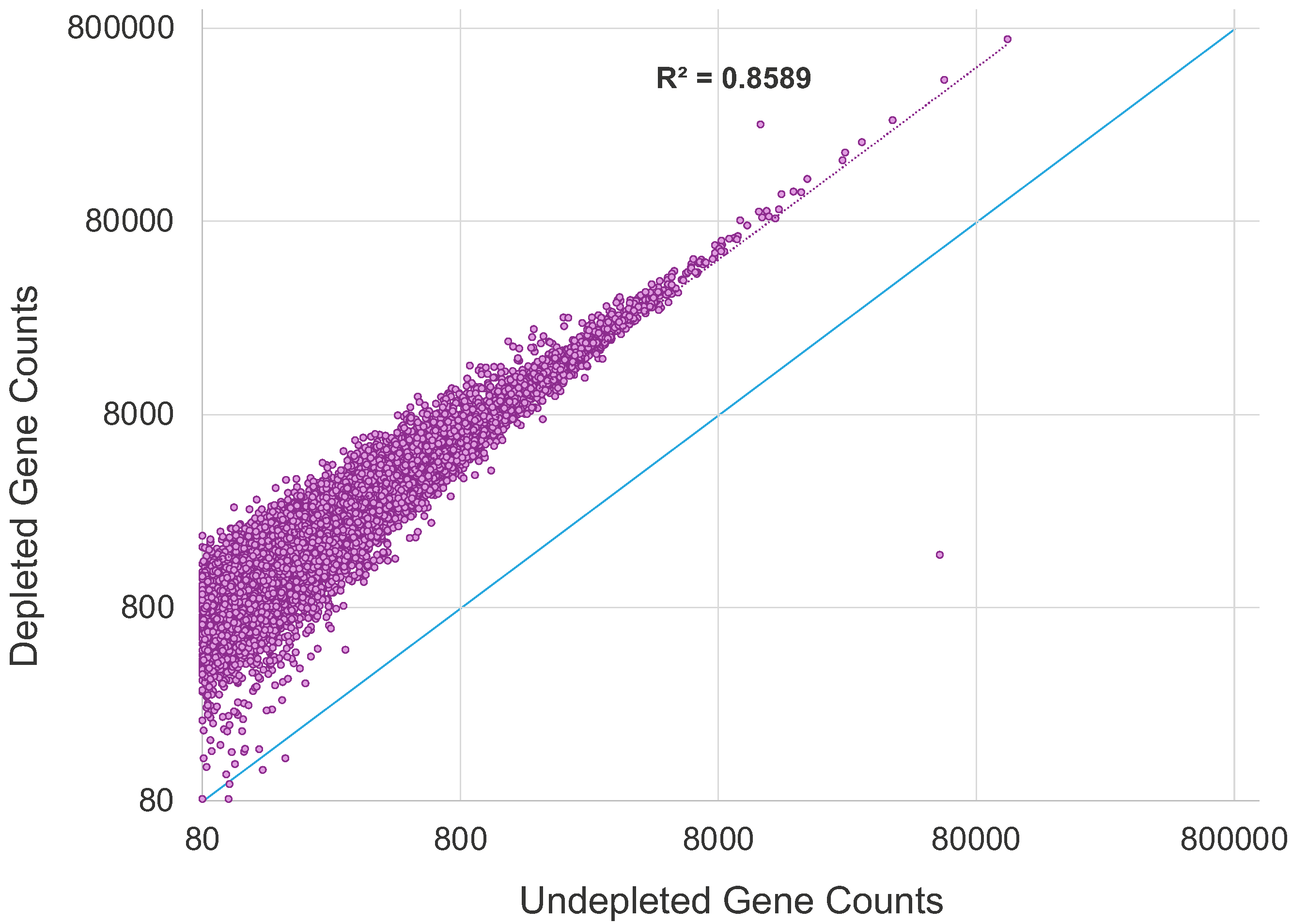
More gene counts in human gene expression
Increased human gene detection sensitivity
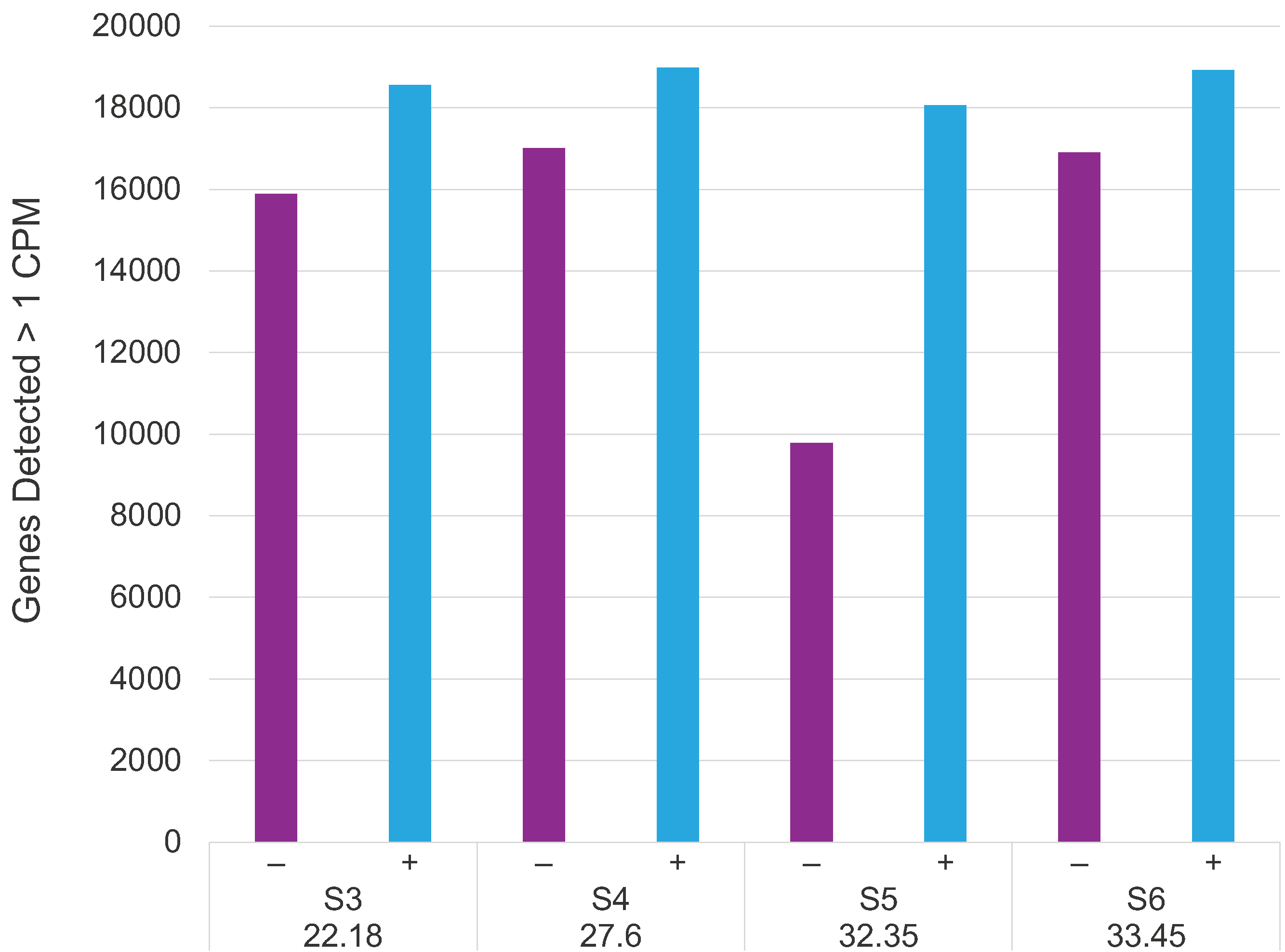
Increased bacterial detection sensitivity
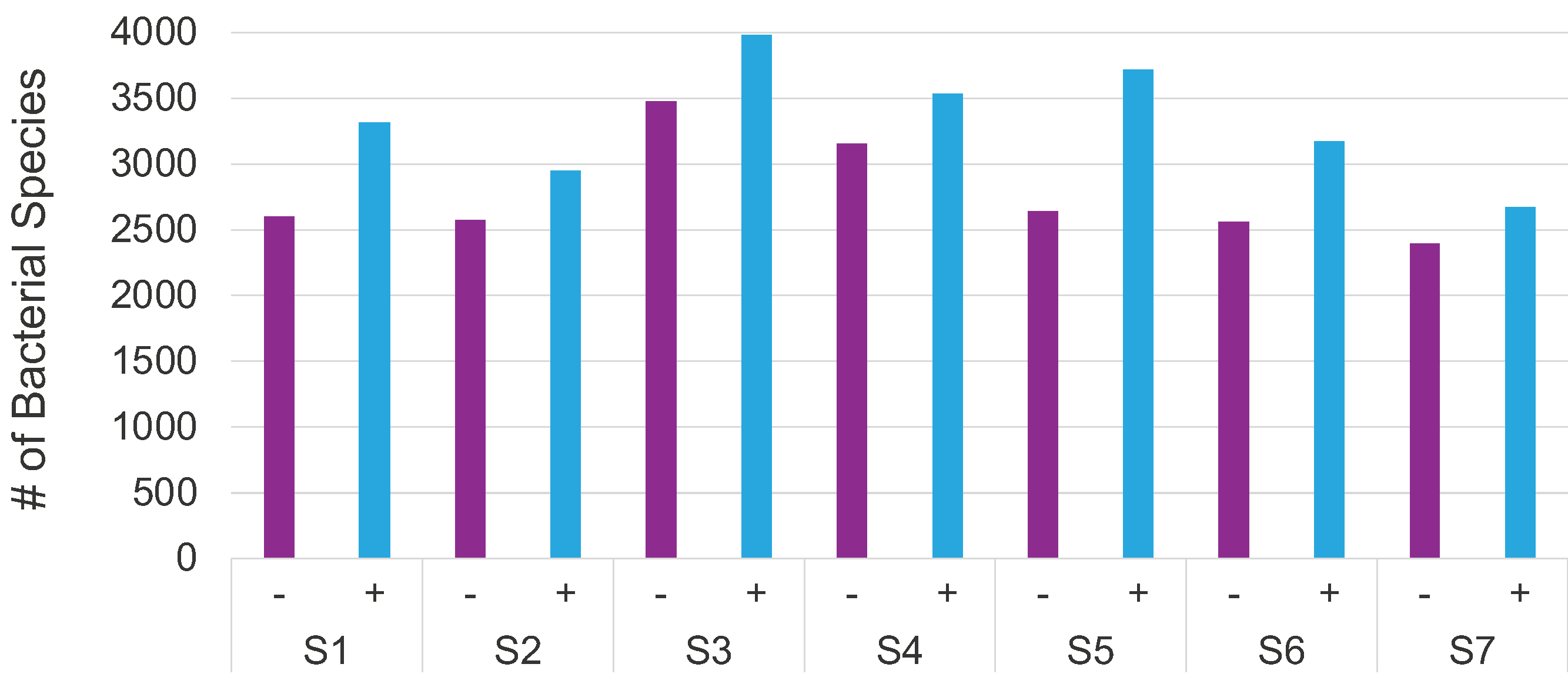

CRISPRclean Plus Stranded Total RNA Prep with rRNA Depletion (Human, Mouse, Rat, Pan Bacteria)
24 samples
$3,600 Add to CartHow can we help maximize your RNA-Seq projects?
Resources
Download helpful product documents.
Connect with us to learn more
For the US patent, the patent number is US 10,604,802 entitled Genome Fractioning. The patent publication is available here.
For the EP patent, the patent number is EP3102722 entitled Genome Fractioning. The patent publication is available here.