
Customer Stories
Removing uninformative RNA helps Weatherall Institute Researchers find rare molecules
The MRC Weatherall Institute of Molecular Medicine (MRC WIMM) at the University of Oxford studies molecular and cell biology and advances translational medicine. By understanding how molecular mechanisms in cells can go awry, MRC WIMM gains new insights into disease and possible solutions. Within the Institute, the MRC Human Immunology Unit is studying the biological mechanisms that govern human immune responses to illuminate how the body reacts to infectious diseases, cancer, allergies and autoimmune conditions.
The unit is particularly focused on inflammatory gut conditions, such as inflammatory bowel disease (IBD), examining the molecular switches that get turned on and off to drive good health or disease. The group uses organoids and other approaches to model various gut conditions, taking primary human biopsy samples and culturing the intestinal stem cells to create 3D structures that mimic intestinal epithelium. The lab has embraced single-cell RNA sequencing (scRNA-seq) to better characterize gut tissue, as well as identifying rare cell types and defining their functions.
Computational biologist Agne Antanaviciute, Ph.D., research assistant and PhD student Ana Sousa Gerós and colleagues are trying to characterize a rare intestinal epithelial cell, a subtype of absorptive cells that no one had described before. Early evidence suggests these cells may play a role in gut regulation, which could make them quite important in IBD and other conditions. Because these cells don’t exist in mice, and are difficult to isolate in people, they are incredibly difficult to study. Antanaviciute and Gerós needed to isolate more so they could manipulate them and better understand their function.
“We were trying to optimize conditions that would generate this rare population of cells,” said Gerós. “What we have really needed is a high throughput testing system, which means a lot of sequencing.”
They tried RNA sequencing but had difficulty finding transcriptional data on these cells. The lab eventually succeeded with extensive scRNA-seq. Still, scRNA-seq can produce a lot of data noise from housekeeping RNAs, including ribosomal and (in this case) particularly mitochondrial. This interfered with their ability to collect data, as well as increasing their sequencing costs. To gain the necessary sequencing depth, and collect the data they needed, they turned to Jumpcode’s CRISPRclean Single Cell RNA Boost Kit. The option of having an off-the-shelf CRISPRclean kit that depleted mitochondria, ribosomal and other transcripts that were superfluous for secondary analysis sounded quite promising.
“We decided to use Jumpcode in this experiment because we could remove the sequences we didn’t want but leave a higher diversity of library. The depletion approach leaves us a lot more data, and we’re slashing our sequencing costs by half, allowing us to increase sequencing depth and run more samples.“
Agne Antanaviciute, Ph.D.
Finding a tiny signal
The lab tried a number of approaches to get more data out of these rare gut cells. Targeted panels were a starting point but produce mixed results. When analyzing the data, the team noticed that some of the genes they wanted to investigate were not in the panel. They also tried creating their own CRISPR depletion approaches, focusing on removing mitochondrial RNA, the major contaminant in their assays. This worked reasonably well, but the workflow was both time-consuming and cumbersome. While the starting point was depleting mitochondrial and other housekeeping RNA, the primary goal was increasing sequencing depth, analyzing more samples and boosting overall sensitivity. They needed a better solution and turned to Jumpcode.
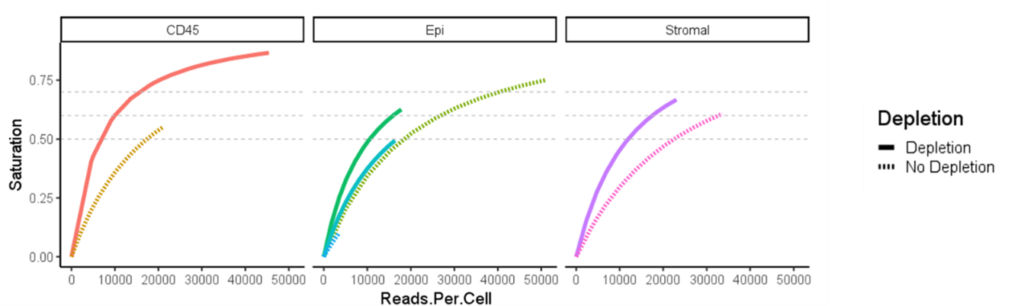
“We decided to use Jumpcode in this experiment because we could remove the sequences we didn’t want but leave a higher diversity of library,” said Dr. Antanaviciute. “The depletion approach leaves us a lot more data, and we’re slashing our sequencing costs by half, allowing us to increase sequencing depth and run more samples.” Depleting unwanted housekeeping RNA provides better opportunities to detect low-prevalence genes, which is essential to learn more about the rare gut cells they are trying to study.
“We detect more genes per cell, and that’s wonderful,” said Dr. Antanaviciute. “We end up with a greater dynamic range and a slightly greater detection rate. It gives us a lot more confidence in our results, and by depleting unwanted RNA, we can actually detect more.”
For the immunology researchers at the Weatherall Institute, Jumpcode provided the complete package, depleting uninformative sequences and helping them gather more data to investigate the rare genes they were looking for with an easy-to-use assay. “Technically it’s a very nice and simple protocol,” said Gerós. “It’s very easy and quick and, technically, quite pleasant as well.”