Enhanced Single Cell RNA-Seq with the BD Rhapsody™ System
Remove mitochondrial interference and enhance single cell RNASeq data through Jumpcode DepleteX® and the BD RhapsodyTM System
Mitochondrial RNA is a big challenge in single cell RNA-Seq studies especially in low viability samples. This contamination can result in unnecessary sequencing of mitochondrial genes that may be undesired. Combining the BD Rhapsody™ Whole Transcriptome Analysis (WTA) Amplification Kit with Jumpcode’s DepleteX assay provides improved data quality that can lead to more valuable biological insights in challenging samples exhibiting high mitochondrial reads.

Highlights
- Compatible with both single cell and single nuclei RNA seq
- Mitochondrial mRNA reduced by 98% in cells and 86% in nuclei samples
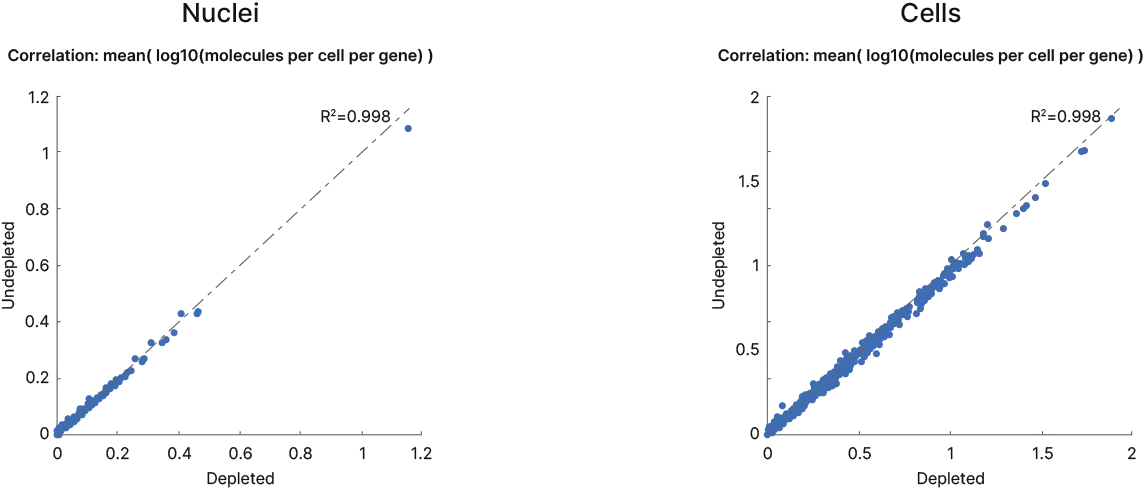
- Biological insights preserved with no significant change in cell numbers, cell composition and gene expression
- Achieved with only 45 minutes of extra hands-on time
Simple workflow, powerful results.
A simple three-step workflow can be integrated into the BD Rhapsody™ WTA protocol to address potential limitations in detecting biologically relevant mRNA expression due to elevated mitochondrial reads in challenging samples
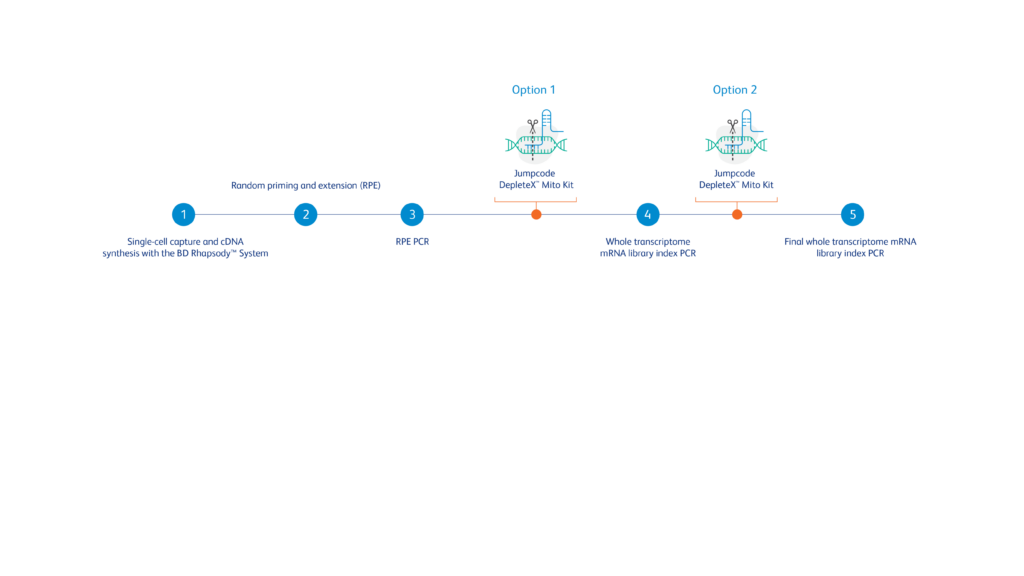
*Data shown here is generated using Depletion option 2.
Jumpcode DepleteX™ effectively removes mitochondrial fragments prior to sequencing
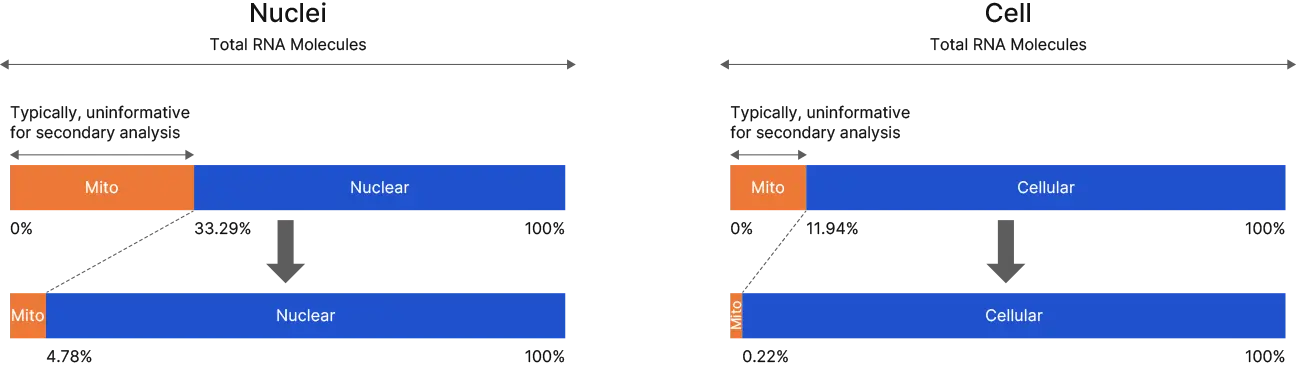
Mitochondrial transcripts comprise between 10-30% of raw reads are reduced substantially to 0.22% and 4.78% after depletion.
Data from PBMCs, some variability expected with different cell types.
Depletion of undesired fragments without biasing the remaining transcriptome