
DepleteX™ Single Cell RNA Boost Kit Product Support
The resources below are designed to help you boost your research with Jumpcode. If you have questions, you can reach us at [email protected].
Getting started

Read this before you start your experiment. This PDF download contains a high-level overview of the process, frequently asked questions, and support contact info in case any issues arise during your experiment or analysis.
There are two versions to choose from:
Analysis guidelines
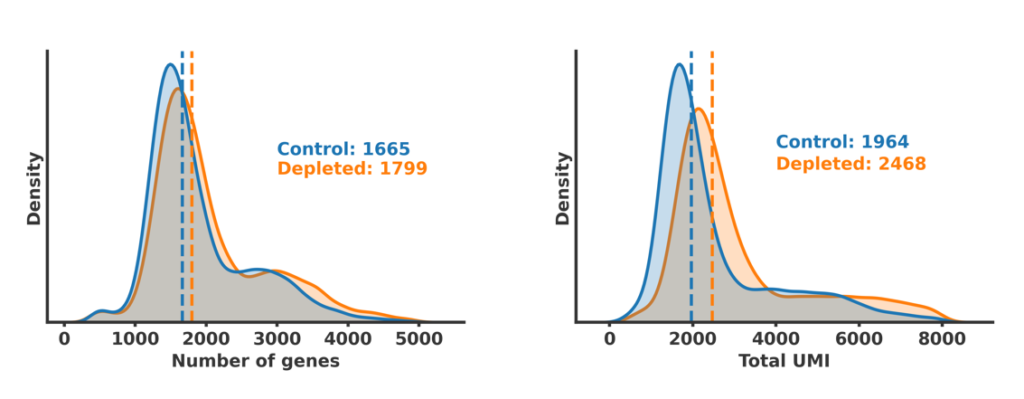
We have multiple options to support your analysis needs. Choose from:
- Video walkthrough with our public dataset files.
- Downsampling guidelines for Cell Ranger
- Python or R step-by-step guide to show you how to analyze your data and see the benefits of depletion
- Single Cell with PacBio MAS-seq github page
Resources
User manual
Depletion content
Download public data set files
FAQs
Can I use the DepleteX™ depletion on mouse samples?
Yes, but the depletion rate is lower since the guides were designed for human samples
Does the DepleteX™ depletion use all of the cDNA from my 10X prep?
No, only ¼ of the available cDNA from a given 10X prep is used.
Is this kit compatible with the 5’ Chromium assay? What about Visium spatial?
Yes, it is compatible with both kits. Our depletion panel is designed to tile across the full length of transcripts.
What is the sequencing depth recommendation for the depleted condition?
In general, single cell users use between 25K –50K reads/cell depending on their application. For the depleted condition, users can choose to sequence 12.5K –25K reads/cell to see the same information or sequence at the same depth to see more information.
Can I use SPRI beads instead of AMPure beads?
Yes, we have tested this in-house and they both work with our assay.